The Acrobat file for the whole document is
HERE.
Transductive learning
In classical machine learning the result of the inference relies
only on the training set and it is always
independent of the test set.
Whilst statistically sound and reasonable, many applications have a
large number of unlabeled input which cannot be used in training, as
for example in chemical or biological areas where there is a plethora
of experimental result but labelling them -- or giving a ``desired
value'' for an experimental outcome is very expensive and
time-consuming (this type of problem is tackled in active
learning).
In
transductive learning the situation, from the machine
learning point of view is simpler, but similar:
all unlabeled points
belong to the test set. This means additional information for the
learning algorithm that needs to be exploited and it can be particularly
important for nonparametric methods: the performance of the sparse
inference using a subset of the training inputs can significantly vary
as a function of the location of the subset (
Basis-,
Support- or
Relevance- Vector set)
on which the approximation is based.
The inclusion of this special knowledge within the GP inference scheme
was recently presented in
Schwaighofer, Tresp
(2003). The inference using this special knowledge is called
query learning and the query points are merely a different
name for the test points.
The methodology proposed here is similar to the above method: the set
of Basis Vectors is fixed a-priori to the set of query points. The GP
is initialised with the query set but no training output data,
accomplished by a constant likelihood. We can then perform
pseudo-learning to learn the locations of the query points -
remember that the associated GP, although having a different
representation, is still the prior process.
Having this modified representation for the prior process, we can
iterate the sparse learning which first projects the GP to the set of
Basis Vectors (which is the query set). The BV set is unaltered during
learning.
In the following a MATLAB code is presented that learns the
two-dimensional sinc function
f(x,y)=sin(r)/r where
r is the radius of the two-dimensional input in polar
coordinates. The function is available within the
OGP package, named
demogp_fixed (uses a fixed BV
set).
Fig.1 below shows the location of the training set (smaller black
'x' signs) and the Basis Vectors which is also the query
set. On Fig.2 one sees the result of the OGP inference using the
query set. Note that due to the (in this case purposeful) mismatch of
model parameters and the function to be approximated, the
approximation is quite poor.
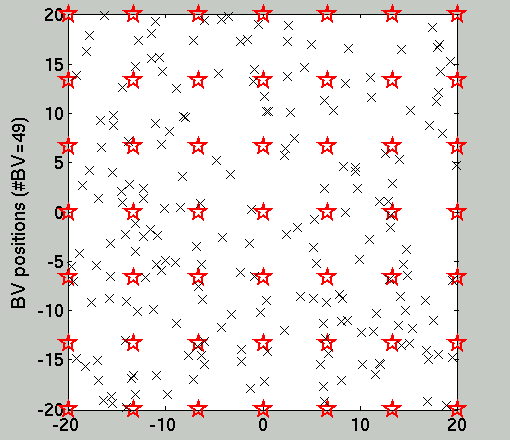
Figure 1. The location of the (200) training
data and the (49) query points.
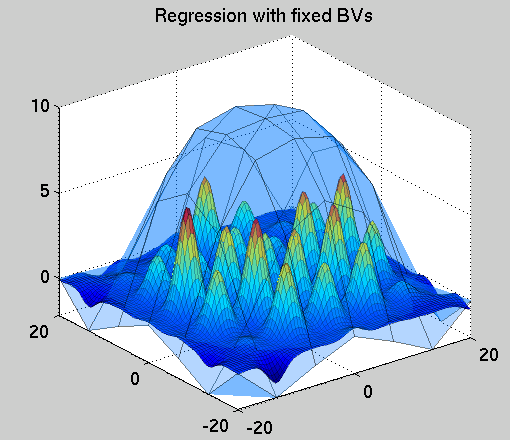
Figure 2. The initial result of the inference using the
predefined set of hyperparameters. It can be seen that the fit
at the training locations is not particularly good, since
the GP cares now only about the query point locations, placed on a
7x7 rectangular grid. The semi-transparent cover is
the function to pe approximated and the surface with the local humps
is the actual approximator.
Further on, as the hyperparameters are better fitted to the function,
the approximation via transductive learning becomes better. This is
visualised on Figs. 3 and 4. The algorithm automatically
detects that the smoothness of the function allows an even sparser
representation and many elements of the query set are removed (they
are redundant), as can be seen from comparing the initial
configuration from Fig. 1 with that of Fig. 5. The Matlab
code is explained in the Section following the
figures.
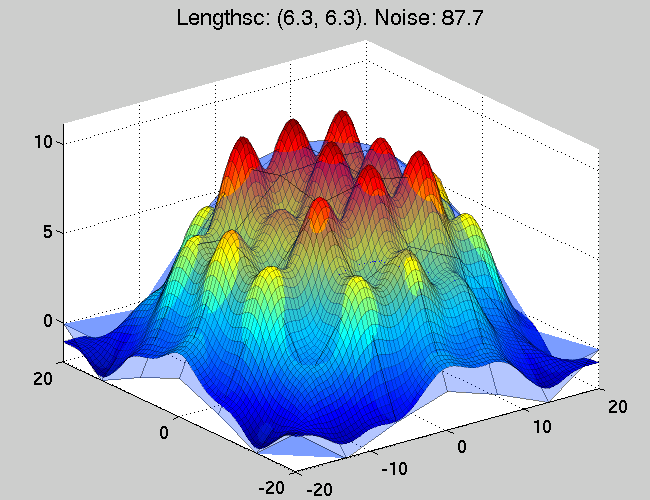
Figure 3. The function approximation after a single
hyper-parameter optimisation step -- an optimisation procedure of
the marginal likelihood. It can be seen that the approximation now
is much better - this is due to the better fit of the GP lengthscale
parameters to the function.
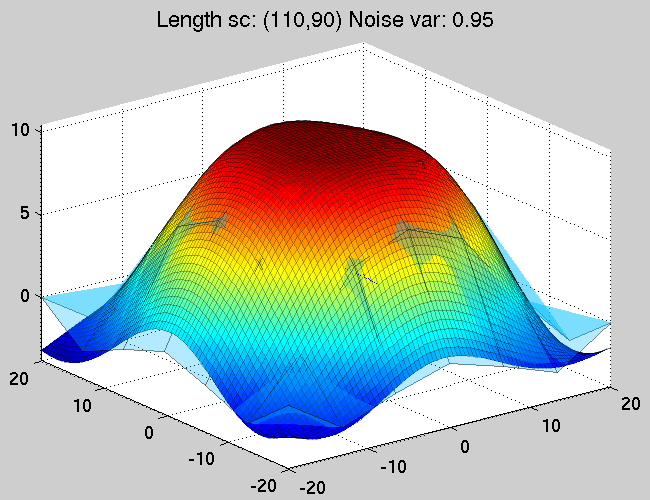
Figure 4. The approximating and the true funtions after
5 steps of iterating between the optimisation of the
GP parameters with fixed hyperparameters and optimising the
hyperparameters whilst keeping the GP parameters fixed.
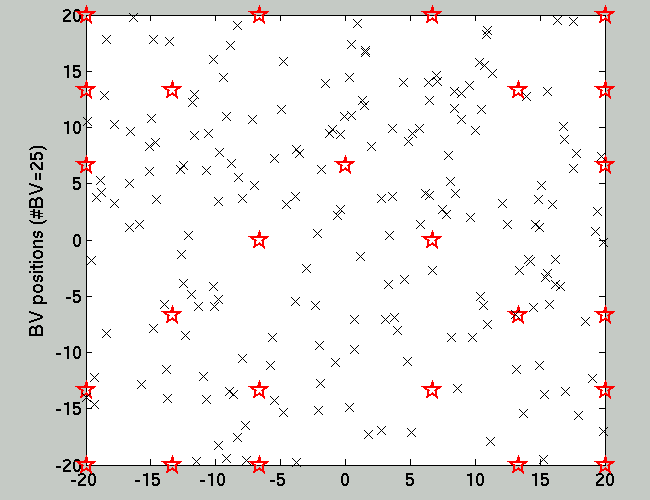
Figure 5. Since the surface of the function to be
approximated is smooth, there are no need for that many Basis
Vectors. The final configuration of the OGP structure includes only
25 elements (sampling or computation with fewer
Basis Vectors is faster). Yet it is clear that the
approximation performance is not affected.
The matlab file, provided in the
code section of these pages,
is named
demogp_fixed.m.
We first declare to global variables we are going to use
global net gpopt ep
The next step is to declare variables related to the data generation
process:
sig02 = 0.3; % noise variance
nType = 'gauss'; % noise TYPE
nTest = 49; % TEST set size = BV SET size
nTrain = 200; % TRAINING set size
gX = [-20:0.5:20];
gN = length(gX);
[pX pY] = meshgrid(gX);
pX = [pX(:) pY(:)];
Then the training and test datasets are generated:
% training points are randomly placed
[xTrain, yTrain] = sinc2data(nType,nTrain,sig02,[],0,20,20);
% test points are on a grid
[xTest, yTest] = sinc2data(nType,nTest,0,[],1,20,20);
We initialise the OGP structure. The first function is similar to a
"constructor", the second function specifies the likelihood we are
using, then the threshold value thresh - for numerical
stability - is set.
ogp(2,1, ... % input/output dimension
'sqexp', ... % kernel type
[1/[2 2] 3 0]); % kernel parameters
ogpinit(@c_reg_gauss,...
0.5, ... % initial value for the lik. par.
@em_gauss); % function to adjust lik. noise
net.thresh = 1e-1; % the admission threshold for new BVs
% SAVING
netS = net;
The next two lines set up the transductive- or query
learning: we initialise the BV-s of the OGP structure
with the test set locations.
% SETTING the initial OGP structure
net = netS;
% FIXED BV set
ogpemptybv(xTest(randperm(size(xTest,1)),:));
net.isBVfixed = 1;
Code that controls the various parameters that will be displayed
during learning (not really important).
% What to print out during learning
gpopt = defoptions; % default values
gpopt.pavg = 1; % log-average
gpopt.ptest = 1; % test error
gpopt.xtest = xTest; % the test inputs
gpopt.ytest = yTest; % desired outputs
gpopt.disperr=0;
gpopt.freq = 10; % frequency of measuring the errors
gpopt.postopt.isep = 1; % USING the TAP/EP algorithm
gpopt.postopt.itn = 3; % number of EP iteration with CHANGING BVs
gpopt.postopt.fixitn= 2; % FIXING the BV set.
The sequence of training and hyper-parameter estimation.
for iHyp=1:5;
ogptrain(xTrain,yTrain);
ogpreset;
fixErr(bInd,:) = gpopt.testerror;
end;
and the rest of the code involves plotting and visualisation (not
explained here, see
demogp_fixed for details)
References
Questions, comments, suggestions: contact Lehel
Csató.





